

Each staining/channel (red, green, blue) can be toggled on or off to allow close examination of the relationships between them. For one of the four data points, the researcher displayed a table of all measurements in the database (c), the raw image (d), or with outlines overlaid (e). To examine the cell cycle distribution of these samples, a DNA content histogram based on individual cells within those four images was plotted (b). The data points highlighted in blue in the scatterplot (a) represent replicates of a particular treatment condition with high mean cell area and nuclear area. The "ImageNumber" and "well" columns provide additional information about the samples the researcher is investigating.ĭata points can be investigated. The first two columns of the table show other information about those data points based on the axes of the scatterplot shown in (a). Finally, the sample names (d, "Hairpin" column, for this particular RNA interference experiment) corresponding to the selected blue data points can be displayed in a table to see which samples are present in the selected points. In this case, the selected blue data points indeed have an unusual cell cycle distribution (fewer 2N cells relative to 4N cells) as compared to all cells in the experiment. As well, a DNA content histogram (c) shows individual cell data from the selected image data points (blue), relative to all cells in the experiment (red). Immediately, the corresponding points appear blue in the other open scatterplot (b), allowing the relationship between all of the plotted features to be examined. Data points representing images with high nuclear area (averaged over the cells in the image) and high DNA intensity (averaged over the cells in the image) are highlighted in blue by brushing the scatterplot shown in (a). Relationships among data can be explored. From this plot, it is apparent that these four data points have high cell and nuclear area (two left-most coordinates) but low cell count (right-most coordinate). The four selected blue data points in the scatterplot are also highlighted in the parallel coordinate plot as blue lines. (d) Parallel coordinate plot where 6 features are plotted, one on each numbered axis as labeled in the table shown below the plot. (c) Density plot of individual cell data (X-axis = area of the cell Y-axis = area of the nucleus. One particular sample and its replicates are highlighted as blue data points, and the blue lines indicate +/- two standard deviations from the mean, although because the data is non-Gaussian, the left standard deviation line is not visible. (b) Scatterplot of image data (X-axis = mean area of all cells in the image Y-axis = mean area of all nuclei in the image).
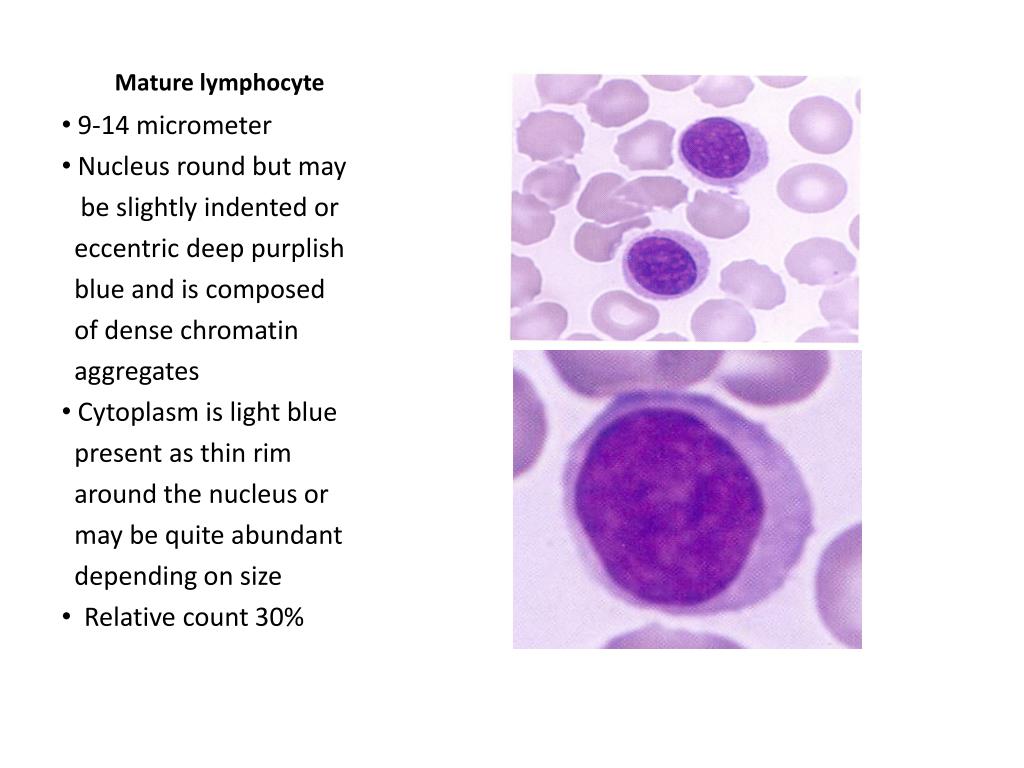
(a) A histogram of per-image data (the mean area of all cells in the image). Four types of plots created by CellProfiler Analyst are shown.


 0 kommentar(er)
0 kommentar(er)
